SciJava Ops from Python
This example demonstrates how to use SciJava Ops with Python. Using SciJava Ops framework with Python depends on scyjava to provide robust
Java code access and imglyb to bridge the ImgLib2 and NumPy data structures. The Python script in this example downloads a 3D 3T3 cell
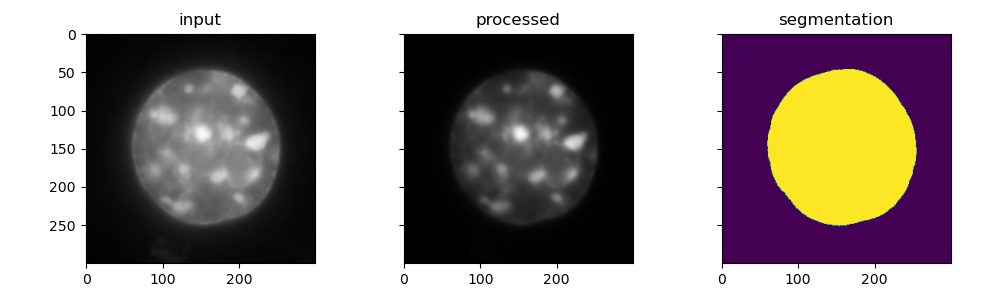
nucleus dataset (with shape: 37, 300, 300), performes image processing with to improve the nucleus signal, segments the nucleus and measures
the 3D volume of the nucleus by creating a mesh. Finally the input image, processed image and the segmented label images are displayed in
matplotlib, and the volume (μm3) is printed to the console.
[INFO]: Adding SciJava repo...
[INFO]: Adding endpoints...
[INFO]: Adding classes...
[INFO]: volume = 456.0139675000041 μm^3
To run this example, create a conda/mamba environment with the following environment.yml file:
name: scijava-ops
channels:
- conda-forge
- defaults
dependencies:
- scyjava
- imglyb
- tifffile
- matplotlib
- requests
- openjdk >= 17
Activate the scijava-ops conda/mamba environment and run the following Python script:
import io
import requests
import imglyb
import scyjava as sj
import numpy as np
import tifffile as tf
import matplotlib.pyplot as plt
from typing import List
def imglib_to_numpy(rai: "net.imglib2.RandomAccessibleInterval", dtype: str) -> np.ndarray:
"""Convert an ImgLib2 image to NumPy.
:param rai: Input RandomAccessibleInterval (RAI)
:param dtype: dtype for output NumPy array
:return: A NumPy array with the specified dtype and data
"""
# create empty NumPy array
shape = list(rai.dimensionsAsLongArray())
shape.reverse() # XY -> row, col
narr = np.zeros(shape, dtype=dtype)
# create RAI reference with imglyb and copy data
ImgUtil.copy(rai, imglyb.to_imglib(narr))
return narr
def numpy_to_imglib(narr: np.ndarray) -> "net.imglib2.RandomAccessibleInterval":
"""Convert a NumPy image to ImgLib2.
:param narr: Input NumPy array
:return: A ImgLib2 RandomAccessibleInterval (reference)
"""
return imglyb.to_imglib(narr)
def read_image_from_url(url: str) -> np.ndarray:
"""Read a .tif image from a URL.
:param url: URL of .tif image
:return: NumPy array of image in URL
"""
return tf.imread(io.BytesIO(requests.get(url).content))
def segment_nuclei(rai: "net.imglib2.RandomAccessibleInterval") -> List:
"""Segment nuclei using SciJava Ops!
:param rai: Input RandomAccessibleInterval (RAI)
:return: A list containing:
(1) Image processing result
(2) Threshold boolean mask
(3) ImgLabeling
"""
# create image containers
mul_result = ops.op("create.img").input(rai, FloatType()).apply()
thres_mask = ops.op("create.img").input(rai, BitType()).apply()
# process image and create ImgLabeling
mean_blur = ops.op("filter.mean").input(rai, HyperSphereShape(5)).apply()
ops.op("math.mul").input(rai, mean_blur).output(mul_result).compute()
ops.op("threshold.huang").input(mul_result).output(thres_mask).compute()
labeling = ops.op("labeling.cca").input(thres_mask, StructuringElement.EIGHT_CONNECTED).apply()
return [mul_result, thres_mask, labeling]
def measure_volume(rai: "net.imglib2.RandomAccessibleInterval", cal: List[float]) -> float:
"""Create a mesh and measure its volume.
:param rai: Input RandomAccessibleInterval (RAI)
:param cal: imaging calibration, with one float per dimension in the input,
in microns
:return: Volume of the 3D mesh
"""
mesh = ops.op("geom.marchingCubes").input(rai).apply()
# Mesh volume returned in voxels
volume = ops.op("geom.size").input(mesh).apply().getRealDouble()
# Convert voxels to um^3
for c in cal:
volume *= c
return volume
# add SciJava repository
print("[INFO]: Adding SciJava repo...")
sj.config.add_repositories({'scijava.public': 'https://maven.scijava.org/content/groups/public'})
# add endpoints
print("[INFO]: Adding endpoints...")
sj.config.endpoints = ['net.imglib2:imglib2',
'net.imglib2:imglib2-imglyb',
'io.scif:scifio',
'org.scijava:scijava-ops-engine:0-SNAPSHOT',
'org.scijava:scijava-ops-image:0-SNAPSHOT']
# import Java classes
print("[INFO]: Adding classes...")
OpEnvironment = sj.jimport('org.scijava.ops.api.OpEnvironment')
BitType = sj.jimport('net.imglib2.type.logic.BitType')
FloatType = sj.jimport('net.imglib2.type.numeric.real.FloatType')
HyperSphereShape = sj.jimport('net.imglib2.algorithm.neighborhood.HyperSphereShape')
ImgUtil = sj.jimport('net.imglib2.util.ImgUtil')
StructuringElement = sj.jimport('net.imglib2.algorithm.labeling.ConnectedComponents.StructuringElement')
# build OpEnvironment
ops = OpEnvironment.build()
# open image
narr = read_image_from_url("https://media.scijava.org/scijava-ops/1.0.0/3t3_nucleus.tif")
cal = [0.065, 0.065, 0.1] # microns, from imaging parameters
rai = numpy_to_imglib(narr)
results = segment_nuclei(rai)
print(f"[INFO]: volume = {measure_volume(results[1], cal)} μm^3")
# display results with matplotlib
processed = imglib_to_numpy(results[0], "float32")
labels = imglib_to_numpy(results[2].getIndexImg(), "int32")
fig, ax = plt.subplots(nrows=1, ncols=3, figsize=(10, 3), sharex=True, sharey=True)
ax[0].imshow(narr[20, :, :], cmap='gray')
ax[0].set_title("input")
ax[1].imshow(processed[20, :, :], cmap='gray')
ax[1].set_title("processed")
ax[2].imshow(labels[20, :, :])
ax[2].set_title("segmentation")
plt.tight_layout()
plt.show()