Spatially Adaptive Colocalization Analysis
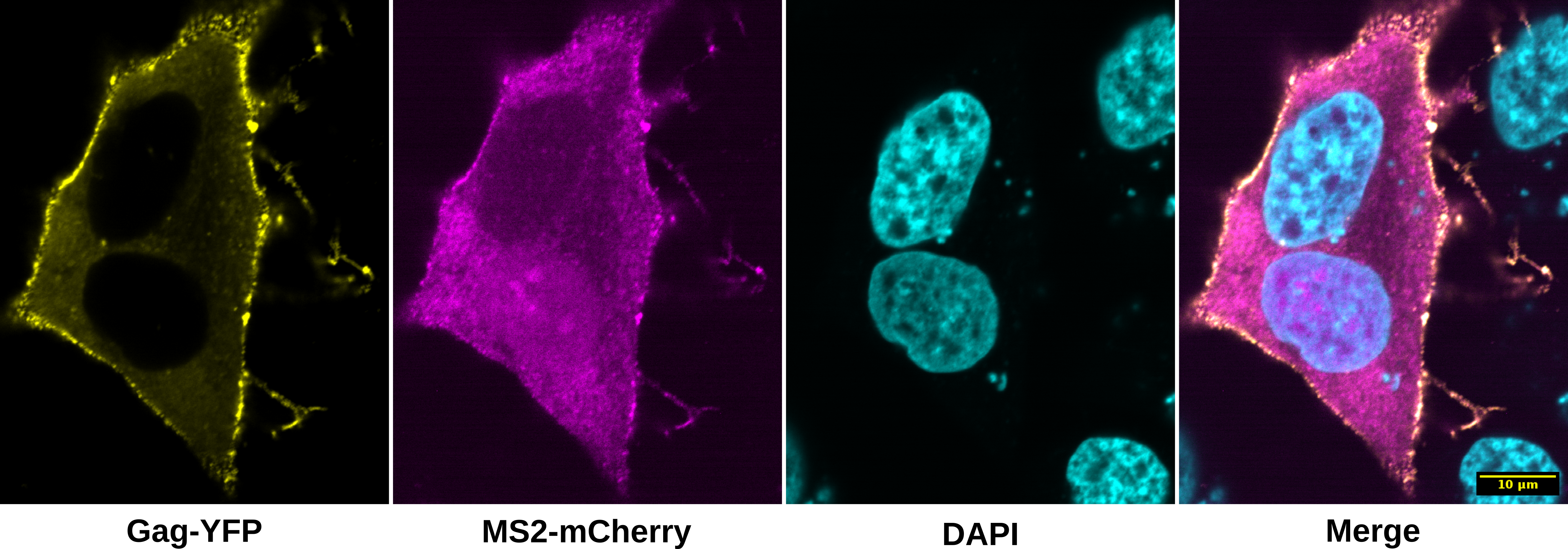
In this example we will use SciJava Ops and the Spatially Adaptive Colocalization Analysis (SACA) 1 framework on HeLa cells transfected with a dual fluorescent HIV-1 NL4-3 construct to colocalize viral mRNAs with HIV-1 NL4-3 Gag proteins. Two fluorescent fusion proteins are made when cells express the modified HIV-1 construct: Gag-mVenus and MS2-mCherry. The Gag-mVenus fusion protein tracks Gag protein, the primary structural component of the HIV-1 virion. The MS2-mCherry fusion protein binds to 24 copies of the MS2 bacteriophage RNA stem-loop 2 inserted into the HIV-1 construct, enabling fixed and live-cell imaging of viral mRNA dynamics. Taken together, this system tracks both Gag and viral mRNAs from the cell’s cytoplasm to sites of viral particle assembly at the plasma membrane where they colocalize 3. This example uses fixed cell data collected with a laser-scanning confocal microscope at 1.4NA 60x magnification (oil immersion).
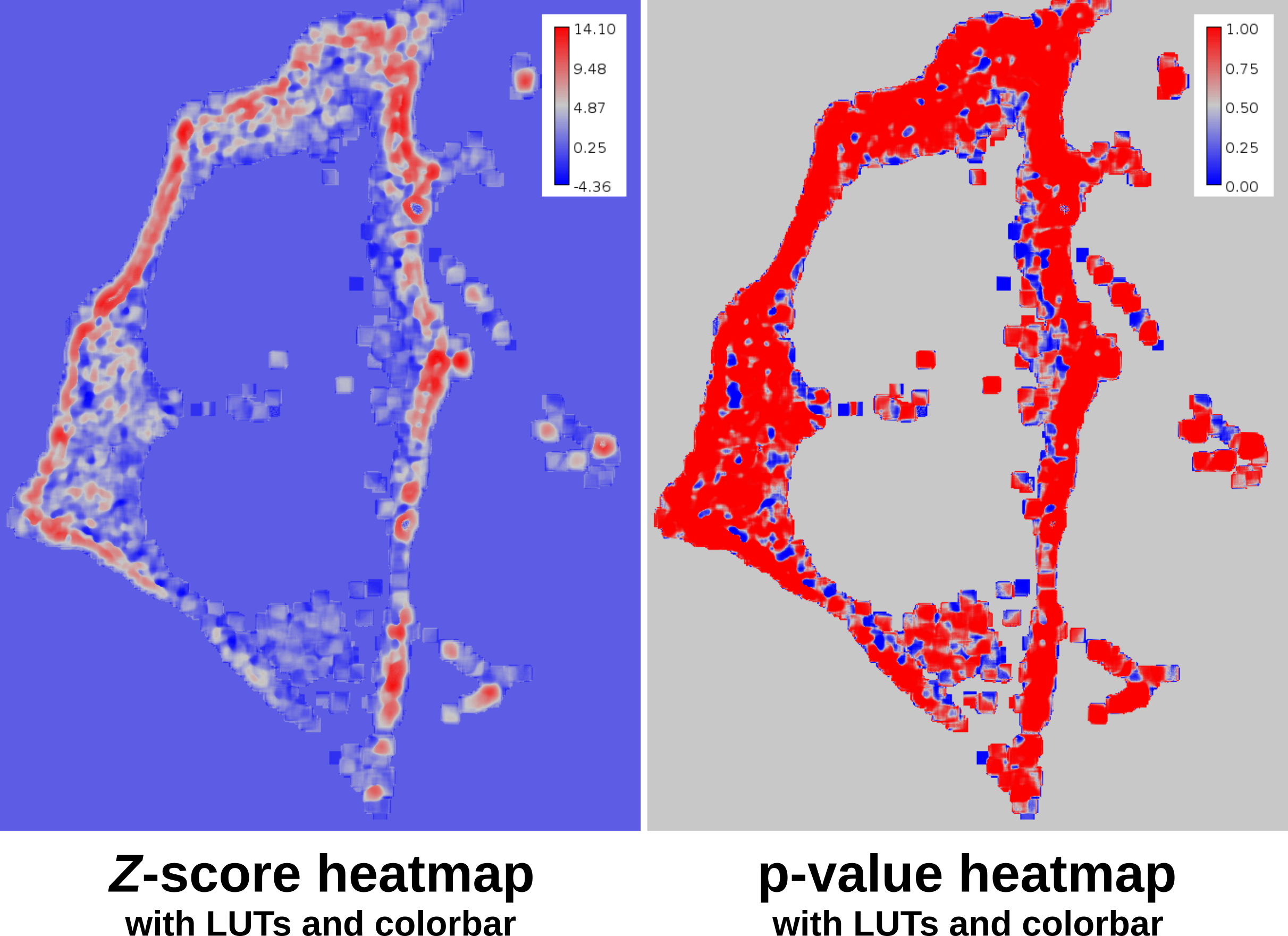
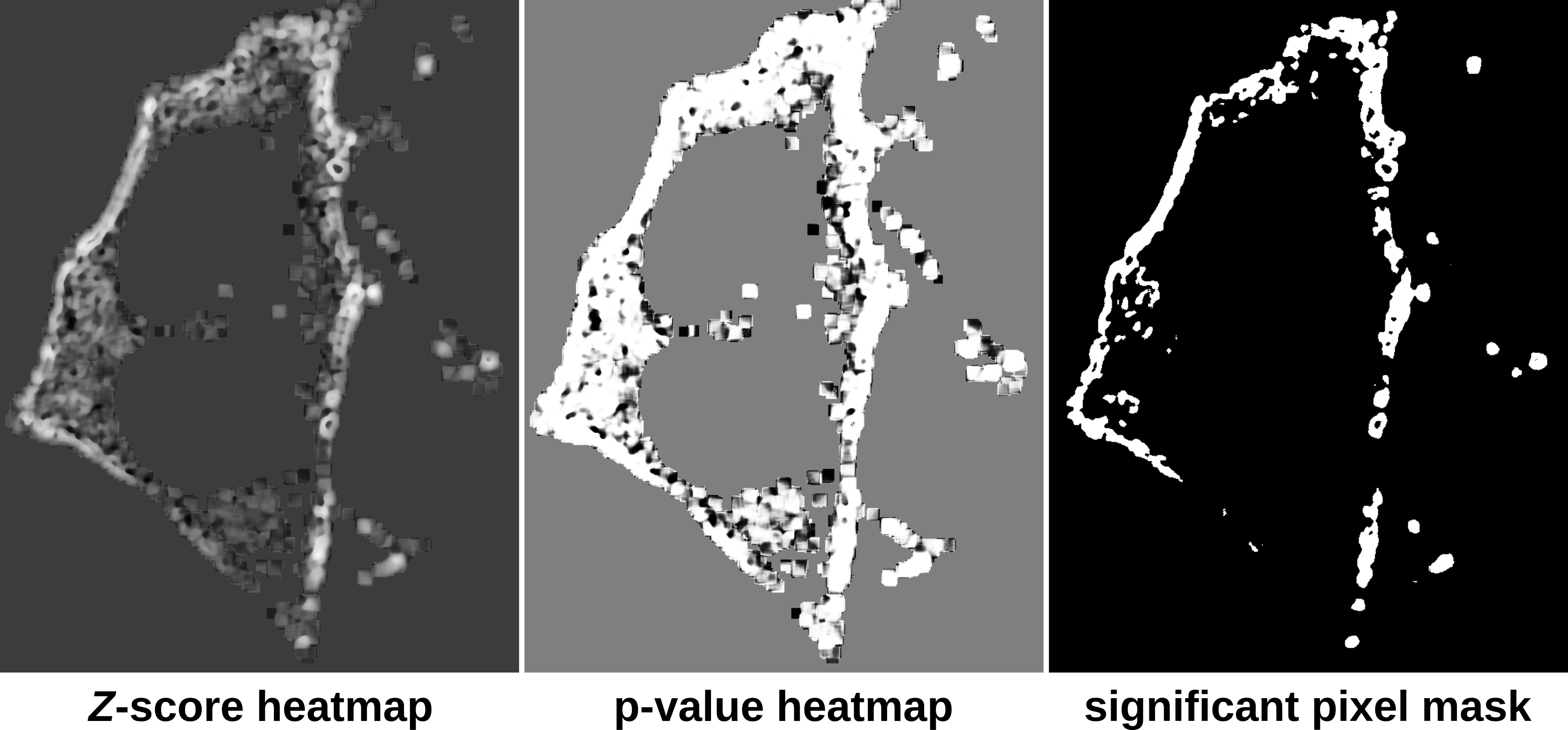
The SACA framework produces two outputs: a Z-score heatmap and a significant pixel mask. The Z-score heatmap
indicates the colocalization or anti-colocalization strength at a pixel-wise level. A pixel-wise p-value of the colocalization strength
can be computed easily by using the Z-score heatmap with the stats.pnorm Op. The significant pixel mask identifies which pixels are
significantly colocalized. This example script takes advantage of this feature of the SACA framework and utilizes the significant pixel
mask as a region of interest to compute Pearson’s 4 and Li’s 5 colocalization coefficients.
You can download the colocalization dataset here.
SciJava Ops via Fiji’s scripting engine with script parameters:
#@ OpEnvironment ops
#@ ConvertService cs
#@ Img input
#@output zscore
#@output pvalue
#@output sig_mask
import net.imglib2.type.logic.BitType
import net.imglib2.roi.labeling.ImgLabeling
import net.imglib2.roi.labeling.LabelRegions
import net.imglib2.roi.Regions
// split input image into channels
channels = []
input.dimensionsAsLongArray()[2].times { i ->
channels.add(ops.op("transform.hyperSliceView").input(input, 2, i).apply())
}
// create SACA Z-score heatmap
zscore = ops.op("coloc.saca.heatmapZScore").input(channels[0], channels[1]).apply()
// compute pixel-wise p-value
pvalue = ops.op("stats.pnorm").input(zscore).apply()
// create SACA significant pixel mask
sig_mask = ops.op("create.img").input(channels[0], new BitType()).apply()
ops.op("coloc.saca.sigMask").input(zscore).output(sig_mask).compute()
// convert SACA sig mask into labeling and run
// Pearson's and Li's colocalization quotients
labeling = cs.convert(sig_mask, ImgLabeling)
regs = new LabelRegions(labeling)
coloc_region = regs.getLabelRegion(1)
subsample_1 = Regions.sample(coloc_region, channels[0])
subsample_2 = Regions.sample(coloc_region, channels[1])
pearsons = ops.op("coloc.pearsons").input(subsample_1, subsample_2).apply()
li = ops.op("coloc.icq").input(subsample_1, subsample_2).apply()
// print Pearson's and Li's results
print("Pearson's: " + pearsons + "\nLi's: " + li)
#@ OpEnvironment ops
#@ ConvertService cs
#@ Img input
#@output zscore
#@output pvalue
#@output sig_mask
from net.imglib2.type.logic import BitType
from net.imglib2.roi.labeling import ImgLabeling, LabelRegions
from net.imglib2.roi import Regions
# split input image into channels
channels = []
for i in range(input.dimensionsAsLongArray()[2]):
channels.append(ops.op("transform.hyperSliceView").input(input, 2, i).apply())
# create SACA Z-score heatmap
zscore = ops.op("coloc.saca.heatmapZScore").input(channels[0], channels[1]).apply()
# compute pixel-wise p-value
pvalue = ops.op("stats.pnorm").input(zscore).apply()
# create SACA significant pixel mask
sig_mask = ops.op("create.img").input(channels[0], BitType()).apply()
ops.op("coloc.saca.sigMask").input(zscore).output(sig_mask).compute()
# convert SACA sig mask into labeling and run
# Pearson's and Li's colocalization quotients
labeling = cs.convert(sig_mask, ImgLabeling)
regs = LabelRegions(labeling)
coloc_region = regs.getLabelRegion(1)
subsample_1 = Regions.sample(coloc_region, channels[0])
subsample_2 = Regions.sample(coloc_region, channels[1])
pearsons = ops.op("coloc.pearsons").input(subsample_1, subsample_2).apply()
li = ops.op("coloc.icq").input(subsample_1, subsample_2).apply()
# print Pearson's and Li's results
print("Pearson's: " + str(pearsons))
print("Li's: " + str(li))
Once the script completes, three gray scale images will be displayed: zscore, pvalue and sig_mask.
Additionally the console will print the Pearson’s and Li’s colocalization coefficients using the significant pixel
mask created from SACA.
Pearson's: 0.65593660643
Li's: 0.211457241276
To apply the phase LUT and a colorbar use the following script and select the input images.
#@ ImagePlus zscore_imp (label="Z-score heatmap")
#@ ImagePlus pvalue_imp (label="p-value heatmap")
import ij.IJ
// apply phase LUT to input images
IJ.run(zscore_imp, "phase", "")
IJ.run(pvalue_imp, "phase", "")
// apply color bar to images
IJ.run(zscore_imp, "Calibration Bar...", "location=[Upper Right] fill=White label=Black number=5 decimal=2 font=12 zoom=1.3 overlay")
IJ.run(pvalue_imp, "Calibration Bar...", "location=[Upper Right] fill=White label=Black number=5 decimal=2 font=12 zoom=1.3 overlay")
#@ ImagePlus zscore_imp (label="Z-score heatmap")
#@ ImagePlus pvalue_imp (label="p-value heatmap")
from ij import IJ
# apply phase LUT to input images
IJ.run(zscore_imp, "phase", "")
IJ.run(pvalue_imp, "phase", "")
# apply color bar to images
IJ.run(zscore_imp, "Calibration Bar...", "location=[Upper Right] fill=White label=Black number=5 decimal=2 font=12 zoom=1.3 overlay")
IJ.run(pvalue_imp, "Calibration Bar...", "location=[Upper Right] fill=White label=Black number=5 decimal=2 font=12 zoom=1.3 overlay")